STA 517 3.0 Programming and Statistical Computing with R
Introduction to the tidyverse
Dr Thiyanga Talagala
Today's menu
What is the tidyverse?
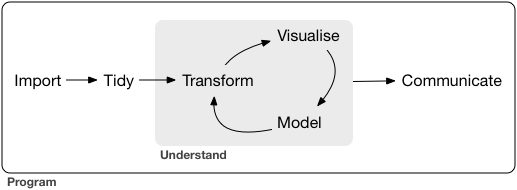
Data science workflow
Tibble
Factor
Pipe

What is the tidyverse?
Collection of essential R packages for data science.
All packages share a common design philosophy, grammar, and data structures.

Setup
install.packages("tidyverse") # install tidyverse packageslibrary(tidyverse) # load tidyverse packages
Workflow: import

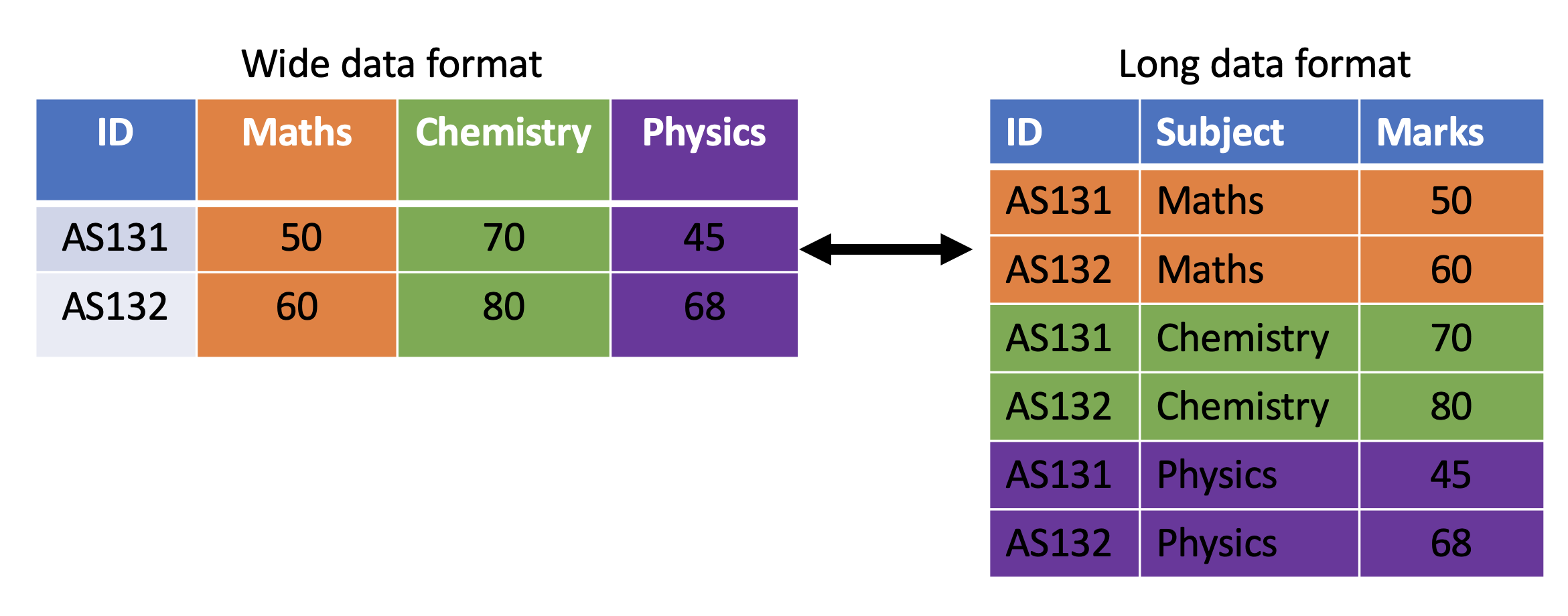
Workflow: tidy

Workflow: transform
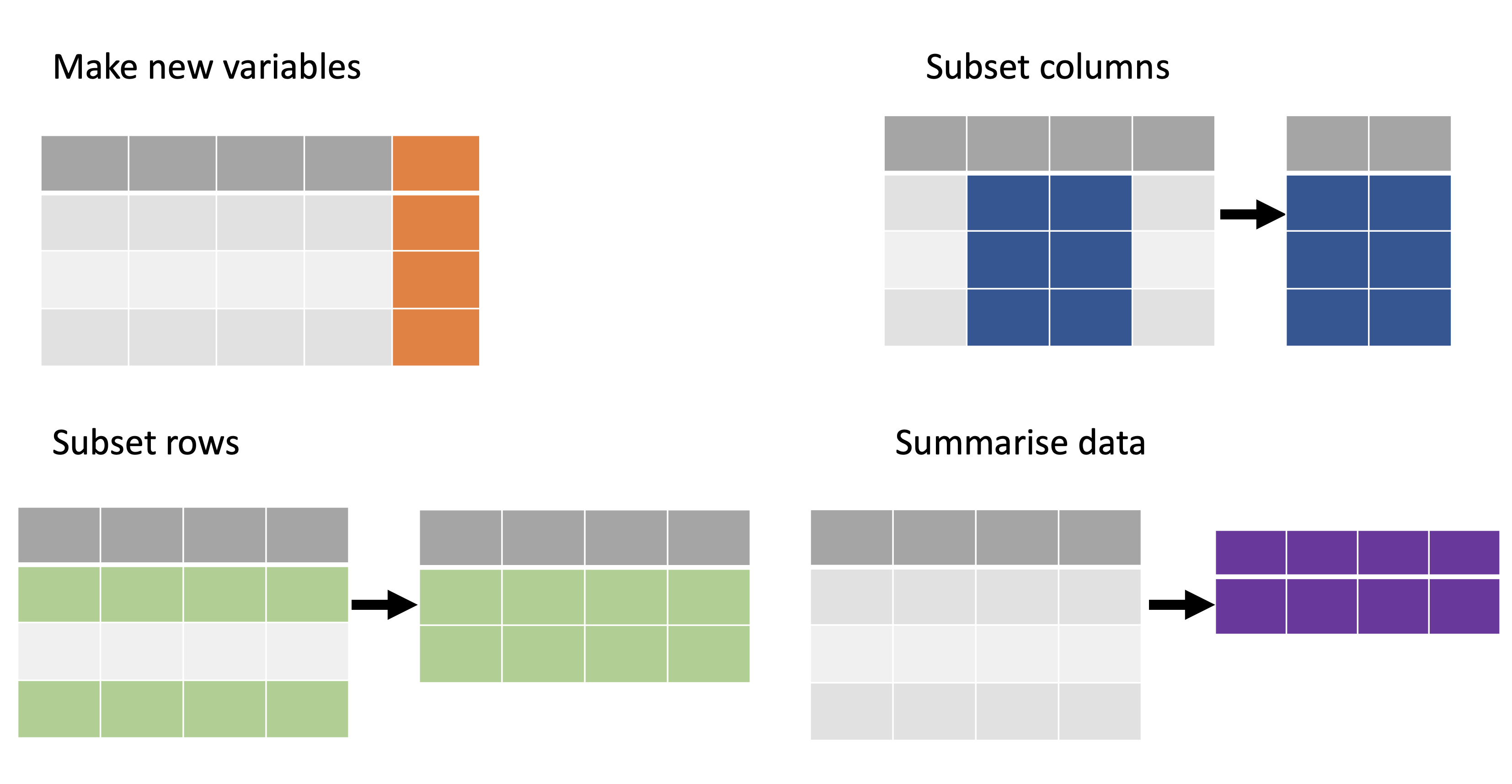

Workflow: visualise
Illustration
library(ggplot2)ggplot(iris, aes(Sepal.Width, Sepal.Length, color=Species)) + geom_point() +theme(aspect.ratio = 1) +scale_color_manual(values = c("#1b9e77", "#d95f02", "#7570b3"))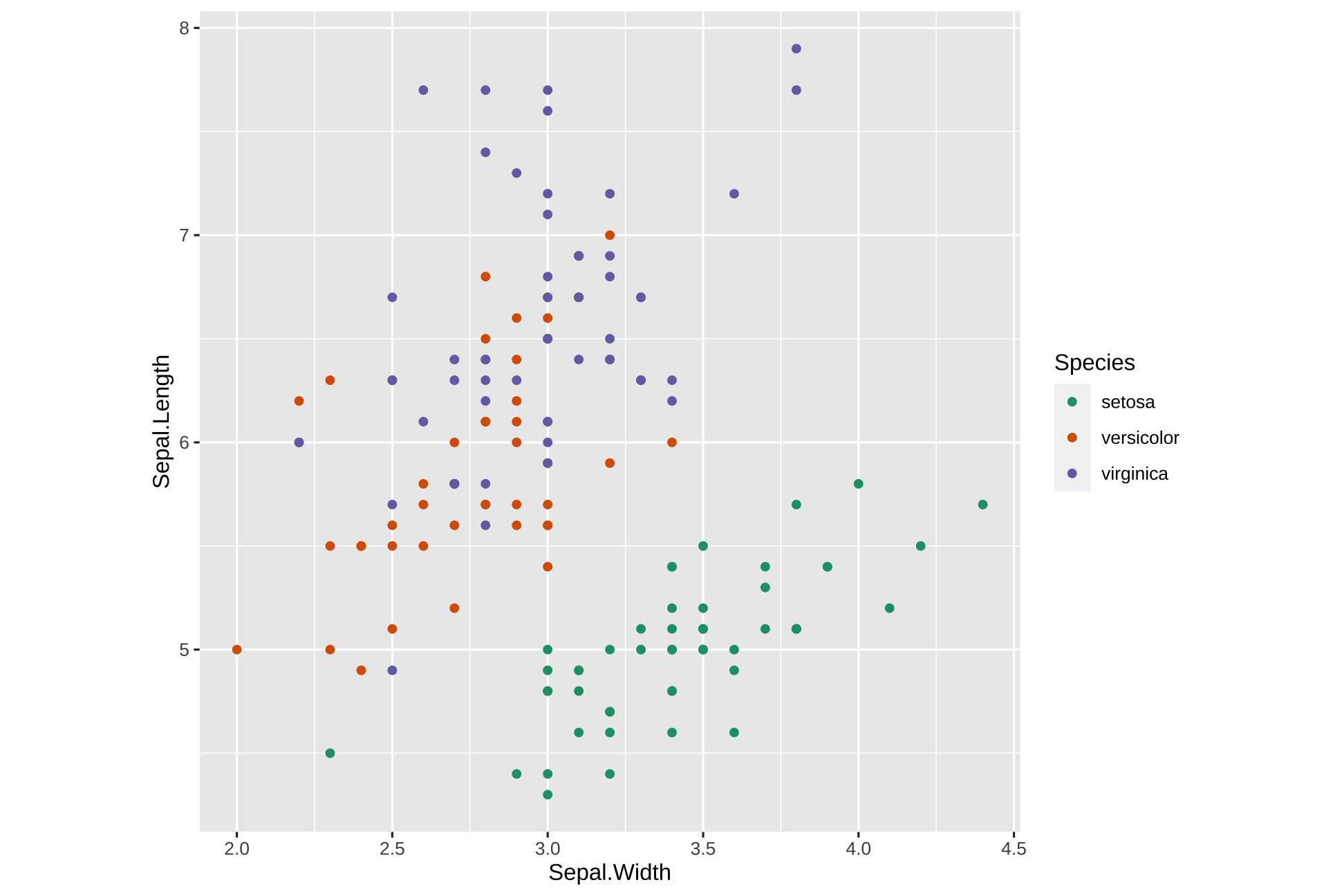
Workflow: model
Illustration: Apply a linear model to each group
nested_iris <- group_by(iris, Species) %>% nest()fit_model <- function(df) lm(Sepal.Length ~ Sepal.Width, data = df)nested_iris <- nested_iris %>% mutate(model = map(data, fit_model))nested_iris$model[[1]] # To print other two models nested_iris$model[[2]] nested_iris$model[[3]]Call:lm(formula = Sepal.Length ~ Sepal.Width, data = df)Coefficients:(Intercept) Sepal.Width 2.6390 0.6905Workflow: communicate

Workflow: communicate
Workflow: R packages
1. Tibble
2. Factor
3. Pipe
Tibble

Tibble
Tibbles are data frames.
A modern re-imagining of data frames.
Create a tibble
library(tidyverse) # library(tibble)first.tbl <- tibble(height = c(150, 200, 160), weight = c(45, 60, 51))first.tbl# A tibble: 3 × 2 height weight <dbl> <dbl>1 150 452 200 603 160 51class(first.tbl)[1] "tbl_df" "tbl" "data.frame"Convert an existing dataframe to a tibble
as_tibble(iris)# A tibble: 150 × 5 Sepal.Length Sepal.Width Petal.Length Petal.Width Species <dbl> <dbl> <dbl> <dbl> <fct> 1 5.1 3.5 1.4 0.2 setosa 2 4.9 3 1.4 0.2 setosa 3 4.7 3.2 1.3 0.2 setosa 4 4.6 3.1 1.5 0.2 setosa 5 5 3.6 1.4 0.2 setosa 6 5.4 3.9 1.7 0.4 setosa 7 4.6 3.4 1.4 0.3 setosa 8 5 3.4 1.5 0.2 setosa 9 4.4 2.9 1.4 0.2 setosa 10 4.9 3.1 1.5 0.1 setosa # … with 140 more rowsConvert a tibble to a dataframe
first.tbl <- tibble(height = c(150, 200, 160), weight = c(45, 60, 51))class(first.tbl)[1] "tbl_df" "tbl" "data.frame"first.tbl.df <- as.data.frame(first.tbl)class(first.tbl.df)[1] "data.frame"tibble vs data.frame
tibble
first.tbl <- tibble(height = c(150, 200, 160), weight = c(45, 60, 51))first.tbl# A tibble: 3 × 2 height weight <dbl> <dbl>1 150 452 200 603 160 51data.frame
dataframe <- data.frame(height = c(150, 200, 160), weight = c(45, 60, 51))dataframe height weight1 150 452 200 603 160 51- You can create new variables that are functions of existing variables.
tibble
first.tbl <- tibble(height = c(150, 200, 160), weight = c(45, 60, 51), bmi = (weight)/height^2)first.tbl# A tibble: 3 × 3 height weight bmi <dbl> <dbl> <dbl>1 150 45 0.002 2 200 60 0.0015 3 160 51 0.00199data.frame
df <- data.frame(height = c(150, 200, 160), weight = c(45, 60, 51), bmi = (weight)/height^2) # Not workingYou will get an error message
Error in data.frame(height = c(150, 200, 160), weight = c(45, 60, 51), :
object 'height' not found.
With data.frame this is how we should create a new variable from the existing columns.
df <- data.frame(height = c(150, 200, 160), weight = c(45, 60, 51)) df$bmi <- (df$weight)/(df$height^2)df height weight bmi1 150 45 0.0020000002 200 60 0.0015000003 160 51 0.001992188- In contrast to data frames, the variable names in tibbles can contain spaces.
Example 1
tbl <- tibble(`patient id` = c(1, 2, 3))tbl# A tibble: 3 × 1 `patient id` <dbl>1 12 23 3df <- data.frame(`patient id` = c(1, 2, 3))df patient.id1 12 23 3- In contrast to data frames, the variable names in tibbles can start with a number.
tbl <- tibble(`1var` = c(1, 2, 3))tbl# A tibble: 3 × 1 `1var` <dbl>1 12 23 3df <- data.frame(`1var` = c(1, 2, 3))df X1var1 12 23 3In general, tibbles do not change the names of input variables and do not use row names.
tibble
A tibble can have columns that are lists.
tbl <- tibble (x = 1:3, y = list(1:3, 1:4, 1:10))tbl# A tibble: 3 × 2 x y <int> <list> 1 1 <int [3]> 2 2 <int [4]> 3 3 <int [10]>data.frame
This feature is not available in data.frame.
If we try to do this with a traditional data frame we get an error.
df <- data.frame(x = 1:3, y = list(1:3, 1:4, 1:10)) ## Not working, errorError in (function (..., row.names = NULL, check.rows = FALSE, check.names = TRUE, : arguments imply differing number of rows: 3, 4, 10
Subsetting: tibble vs data.frame
Subsetting single columns
data.frame
df <- data.frame(x = 1:3, yz = c(10, 20, 30)); df x yz1 1 102 2 203 3 30df[, "x"][1] 1 2 3df[, "x", drop=FALSE] x1 12 23 3tibble
tbl <- tibble(x = 1:3, yz = c(10, 20, 30)); tbl# A tibble: 3 × 2 x yz <int> <dbl>1 1 102 2 203 3 30tbl[, "x"]# A tibble: 3 × 1 x <int>1 12 23 3data.frame
df[, "x"][1] 1 2 3df[, "x", drop=FALSE] x1 12 23 3tibble
tbl[, "x"]# A tibble: 3 × 1 x <int>1 12 23 3Subsetting single columns (cont) - tibble:
tbl <- tibble(x = 1:3, yz = c(10, 20, 30))tbl# A tibble: 3 × 2 x yz <int> <dbl>1 1 102 2 203 3 30tbl[, "x"]# A tibble: 3 × 1 x <int>1 12 23 3# Method 1tbl[, "x", drop = TRUE][1] 1 2 3# Method 2as.data.frame(tbl)[, "x"][1] 1 2 3Subsetting single rows with the drop argument
data.frame
df[1, , drop = TRUE]$x[1] 1$yz[1] 10tibble
tbl[1, , drop = TRUE]# A tibble: 1 × 2 x yz <int> <dbl>1 1 10as.list(tbl[1, ])$x[1] 1$yz[1] 10Accessing non-existent columns
data.frame
df$y[1] 10 20 30df[["y", exact = FALSE]][1] 10 20 30df[["y", exact = TRUE]]NULLtibble
tbl$yNULLtbl[["y", exact = FALSE]]NULLtbl[["y", exact = TRUE]]NULLFunctions work with both tibbles and dataframes
names(), colnames(), rownames(), ncol(), nrow(), length() # length of the underlying listtibble
tb <- tibble(a = 1:3)names(tb)[1] "a"colnames(tb)[1] "a"rownames(tb)[1] "1" "2" "3"nrow(tb); ncol(tb); length(tb)[1] 3[1] 1[1] 1data.frame
df <- data.frame(a = 1:3)names(df)[1] "a"colnames(df)[1] "a"rownames(df)[1] "1" "2" "3"nrow(df); ncol(df); length(df)[1] 3[1] 1[1] 1However, when using tibble, we can use some additional commands
is.tibble(tb)[1] TRUEis_tibble(tb) # is.tibble()` is deprecated as of tibble 2.0.0, Please use `is_tibble()` instead of is.tibble[1] TRUEglimpse(tb)Rows: 3Columns: 1$ a <int> 1, 2, 3Factors
factor
A vector that is used to store categorical variables.
It can only contain predefined values. Hence, factors are useful when you know the possible values a variable may take.
Creating a factor vector
grades <- factor(c("A", "A", "A", "C", "B"))grades[1] A A A C BLevels: A B CNow let's check the class type
class(grades) # It's a factor[1] "factor"Now let's check the class type
class(grades) # It's a factor[1] "factor"To obtain all levels
levels(grades)[1] "A" "B" "C"Creating a factor vector (cont)
- With factors all possible values of the variables can be defined under levels.
grade_factor_vctr <- factor(c("A", "D", "A", "C", "B"), levels = c("A", "B", "C", "D", "E"))grade_factor_vctr[1] A D A C BLevels: A B C D Elevels(grade_factor_vctr)[1] "A" "B" "C" "D" "E"class(levels(grade_factor_vctr))[1] "character"Character vector vs Factor
- Observe the differences in outputs. Factor prints all possible levels of the variable.
Character vector
grade_character_vctr <- c("A", "D", "A", "C", "B")grade_character_vctr[1] "A" "D" "A" "C" "B"Factor vector
grade_factor_vctr <- factor(c("A", "D", "A", "C", "B"), levels = c("A", "B", "C", "D", "E"))grade_factor_vctr[1] A D A C BLevels: A B C D ECharacter vector vs Factor (cont.)
- Factors behave like character vectors but they are actually integers.
Character vector
typeof(grade_character_vctr)[1] "character"Factor vector
typeof(grade_factor_vctr)[1] "integer"- Let's create a contingency table with
tablefunction.
Character vector output with table function
grade_character_vctr <- c("A", "D", "A", "C", "B")table(grade_character_vctr)grade_character_vctrA B C D 2 1 1 1Factor vector (with levels) output with table function
grade_factor_vctr <- factor(c("A", "D", "A", "C", "B"), levels = c("A", "B", "C", "D", "E"))table(grade_factor_vctr)grade_factor_vctrA B C D E 2 1 1 1 0- Output corresponds to factor prints counts for all possible levels of the variable. Hence, with factors it is obvious when some levels contain no observations.
- With factors you can't use values that are not listed in the levels, but with character vectors there is no such restrictions.
Character vector
grade_character_vctr[2] <- "A+"grade_character_vctr[1] "A" "A+" "A" "C" "B"Factor vector
grade_factor_vctr[2] <- "A+"grade_factor_vctr[1] A <NA> A C B Levels: A B C D EModify factor levels
This our factor
grade_factor_vctr[1] A <NA> A C B Levels: A B C D EChange labels
levels(grade_factor_vctr) <- c("Excellent", "Good", "Average", "Poor", "Fail")grade_factor_vctr[1] Excellent <NA> Excellent Average Good Levels: Excellent Good Average Poor FailReverse the level arrangement
levels(grade_factor_vctr) <- rev(levels(grade_factor_vctr))grade_factor_vctr[1] Fail <NA> Fail Average Poor Levels: Fail Poor Average Good ExcellentOrder of factor levels
Default order of levels
fv1 <- factor(c("D","E","E","A", "B", "C"))fv1[1] D E E A B CLevels: A B C D Efv2 <- factor(c("1T","2T","3A","4A", "5A", "6B", "3A"))fv2[1] 1T 2T 3A 4A 5A 6B 3ALevels: 1T 2T 3A 4A 5A 6BOrder of factor levels
Default order of levels
fv1 <- factor(c("D","E","E","A", "B", "C"))fv1[1] D E E A B CLevels: A B C D Efv2 <- factor(c("1T","2T","3A","4A", "5A", "6B", "3A"))fv2[1] 1T 2T 3A 4A 5A 6B 3ALevels: 1T 2T 3A 4A 5A 6Bqplot(fv2, geom = "bar")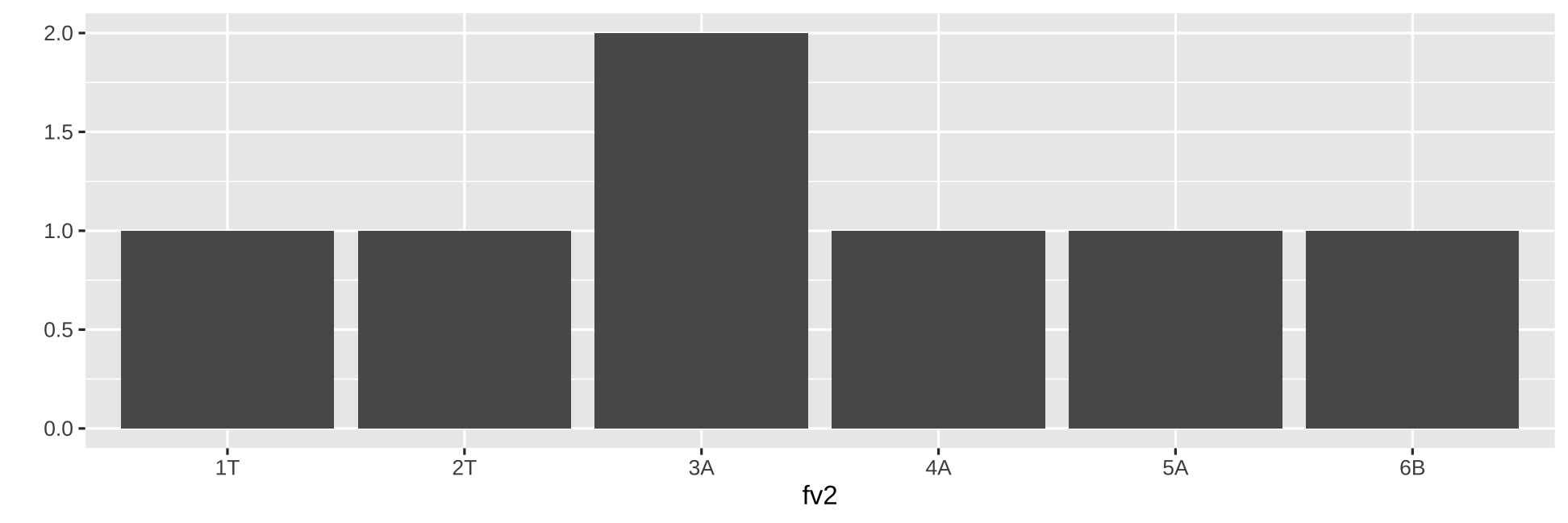
Order of factor levels (cont.)
You can change the order of levels
fv2 <- factor(c("1T","2T","3A","4A", "5A", "6B", "3A"), levels = c("3A", "4A", "5A", "6B", "1T", "2T"))fv2[1] 1T 2T 3A 4A 5A 6B 3ALevels: 3A 4A 5A 6B 1T 2Tqplot(fv2, geom = "bar")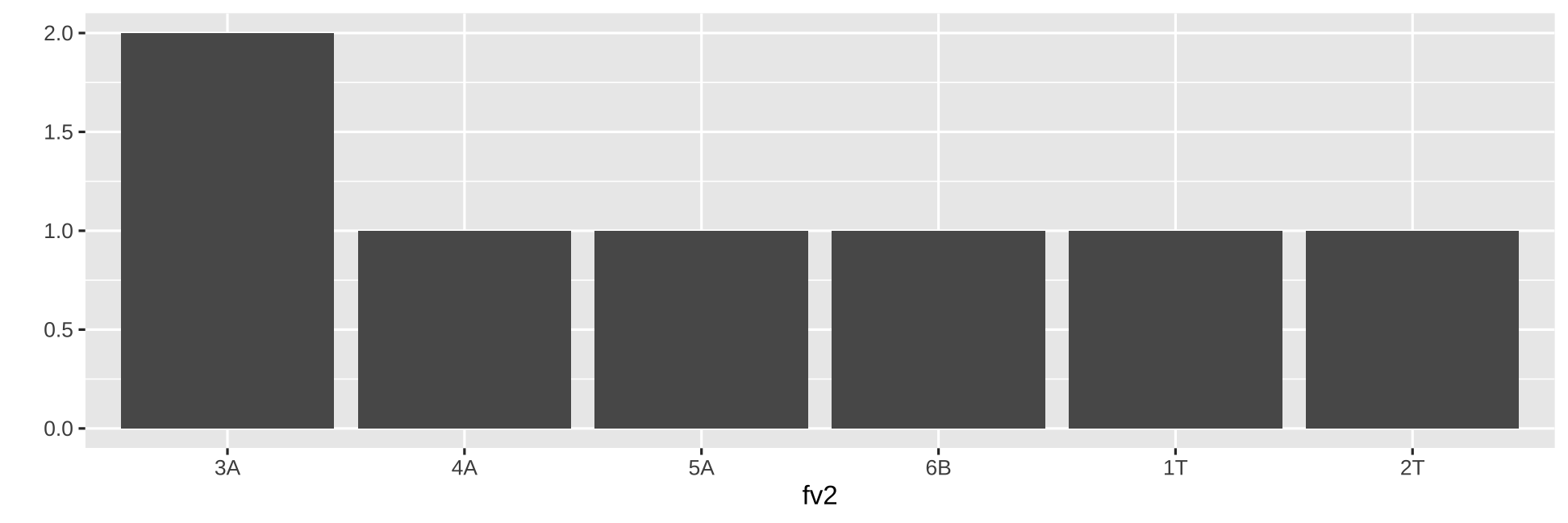
Note that tibbles do not change the types of input variables (e.g., strings are not converted to factors by default).
tbl <- tibble(x1 = c("setosa", "versicolor", "virginica", "setosa"))tbl# A tibble: 4 × 1 x1 <chr> 1 setosa 2 versicolor3 virginica 4 setosadf <- data.frame(x1 = c("setosa", "versicolor", "virginica", "setosa"))df x11 setosa2 versicolor3 virginica4 setosaclass(df$x1)[1] "character"Pipe operator: %>%

Required package: magrittr
install.packages("magrittr")library(magrittr)What does it do?
It takes whatever is on the left-hand-side of the pipe and makes it the first argument of whatever function is on the right-hand-side of the pipe.
For instance,
mean(1:10)[1] 5.5can be written as
1:10 %>% mean()[1] 5.5Pipe operator: %>%
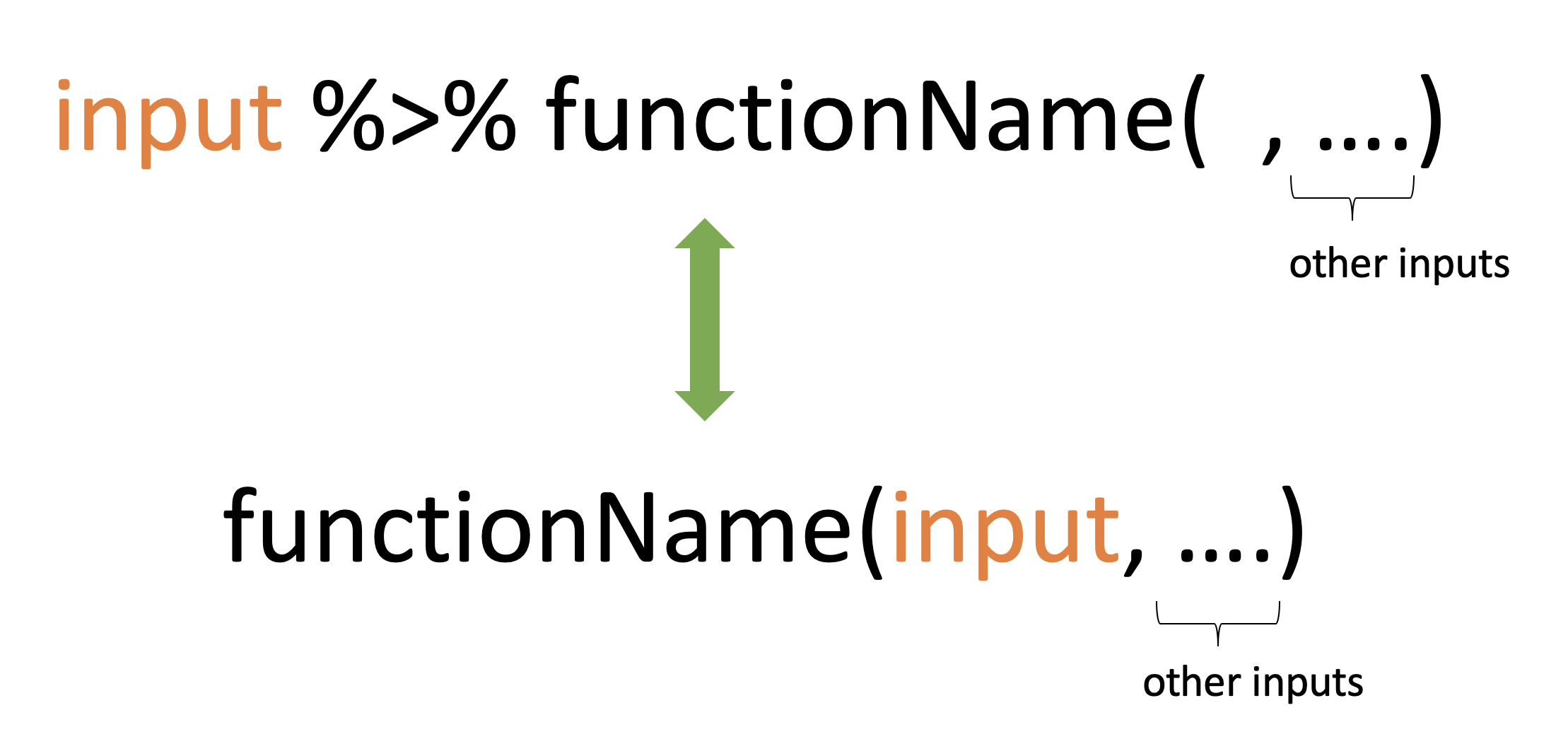
Illustrations
x %>% f(y)turns intof(x, y)x %>% f(y) %>% g(z)turns intog(f(x, y), z)
Why %>%
- This helps to make your code more readable.
Method 1: Without using pipe (hard to read)
colSums(matrix(c(1, 2, 3, 4, 8, 9, 10, 12), nrow=2))[1] 3 7 17 22Method 2: Using pipe (easy to read)
c(1, 2, 3, 4, 8, 9, 10, 12) %>% matrix( , nrow = 2) %>% colSums()[1] 3 7 17 22or
c(1, 2, 3, 4, 8, 9, 10, 12) %>% matrix(nrow = 2) %>% # remove comma colSums()[1] 3 7 17 22library(tidyverse) # to use as_tibblelibrary(magrittr) # to use %>%df <- data.frame(x1 = 1:3, x2 = 4:6)df x1 x21 1 42 2 53 3 6Rules
Rule 1
head(df) df %>% head() x1 x21 1 42 2 53 3 6Rule 2
head(df, n = 2) df %>% head(n = 2) x1 x21 1 42 2 5Rule 3
head(df, n = 2)2 %>% head(df, n = .) x1 x21 1 42 2 5Rule 4
head(as_tibble(df), n = 2)df %>% as_tibble() %>%head(n = 2)# A tibble: 2 × 2 x1 x2 <int> <int>1 1 42 2 5Rule 5: subsetting
df$x1df %>% .$x1[1] 1 2 3or
df[["x1"]]df %>% .[["x1"]][1] 1 2 3or
df[[1]]df %>% .[[1]][1] 1 2 3Offline reading materials
Type the following codes to see more examples:
vignette("magrittr")vignette("tibble")